Code
python==3.8.8
sklearn==1.0.2
numpy==1.20.1
pandas==1.2.3
matplotlib==3.5.1March 4, 2022
sklearn, transformer, pipeline, ColumnTransformer, FunctionTransformer

This notebook shows various ways to work with Skearn Pipelines. * We will start with some of the limitations of pipelines and how to overcome them * We will discuss getting a dataframe from a pipeline instead of a NumPy array, and the benefits of this approach * We will learn how to use CustomTransformer and a FunctionTransformer * We will also build a custom transformer to do some feature engineering * Along the way, we will also see how to avoid common mistakes while creating pipelines
python==3.8.8
sklearn==1.0.2
numpy==1.20.1
pandas==1.2.3
matplotlib==3.5.1For this example we will use the original Titanic dataset, describing the survival status of individual passengers on the Titanic ship.
Some notes from original source: * The variables on our extracted dataset are ‘pclass’, ‘name’, ‘sex’, ‘age’, ‘sibsp’, ‘parch’, ‘ticket’, ‘fare’,‘cabin’, ‘embarked’, ‘boat’, ‘body’, and ‘home.dest’. * pclass refers to passenger class (1st, 2nd, 3rd), and is a proxy for socio-economic class. * Age is in years, and some infants had fractional values. * sibsp = Number of Siblings/Spouses aboard * parch = Number of Parents/Children aboard * The target is either a person survived or not (1 or 0)
Important note: The purpose of this notebook is not to train a best model on titanic data, but to understand the working of Sklearn pipeline and transformers. So please be mindful of that.
from sklearn.datasets import fetch_openml
from sklearn.model_selection import train_test_split
import numpy as np
np.random.seed(42) # for consistency
# Load data from https://www.openml.org/d/40945
X, y = fetch_openml("titanic", version=1, as_frame=True, return_X_y=True)
X.head()| pclass | name | sex | age | sibsp | parch | ticket | fare | cabin | embarked | boat | body | home.dest | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.0 | Allen, Miss. Elisabeth Walton | female | 29.0000 | 0.0 | 0.0 | 24160 | 211.3375 | B5 | S | 2 | NaN | St Louis, MO |
| 1 | 1.0 | Allison, Master. Hudson Trevor | male | 0.9167 | 1.0 | 2.0 | 113781 | 151.5500 | C22 C26 | S | 11 | NaN | Montreal, PQ / Chesterville, ON |
| 2 | 1.0 | Allison, Miss. Helen Loraine | female | 2.0000 | 1.0 | 2.0 | 113781 | 151.5500 | C22 C26 | S | None | NaN | Montreal, PQ / Chesterville, ON |
| 3 | 1.0 | Allison, Mr. Hudson Joshua Creighton | male | 30.0000 | 1.0 | 2.0 | 113781 | 151.5500 | C22 C26 | S | None | 135.0 | Montreal, PQ / Chesterville, ON |
| 4 | 1.0 | Allison, Mrs. Hudson J C (Bessie Waldo Daniels) | female | 25.0000 | 1.0 | 2.0 | 113781 | 151.5500 | C22 C26 | S | None | NaN | Montreal, PQ / Chesterville, ON |
##
# let's check the frequency of missing values in each feature
X.isnull().sum().sort_values(ascending=False)body 1188
cabin 1014
boat 823
home.dest 564
age 263
embarked 2
fare 1
pclass 0
name 0
sex 0
sibsp 0
parch 0
ticket 0
dtype: int64##
# let's drop top 4 features with highest percentage of missing data
# This step is done to make our working with pipeline simpler and easier to understand
X.drop(['body', 'cabin', 'boat', 'home.dest'], axis=1, inplace=True)
X_train, X_test, y_train, y_test = train_test_split(X, y, stratify=y, test_size=0.2)
X_train.head()| pclass | name | sex | age | sibsp | parch | ticket | fare | embarked | |
|---|---|---|---|---|---|---|---|---|---|
| 999 | 3.0 | McCarthy, Miss. Catherine 'Katie' | female | NaN | 0.0 | 0.0 | 383123 | 7.7500 | Q |
| 392 | 2.0 | del Carlo, Mrs. Sebastiano (Argenia Genovesi) | female | 24.0 | 1.0 | 0.0 | SC/PARIS 2167 | 27.7208 | C |
| 628 | 3.0 | Andersson, Miss. Sigrid Elisabeth | female | 11.0 | 4.0 | 2.0 | 347082 | 31.2750 | S |
| 1165 | 3.0 | Saad, Mr. Khalil | male | 25.0 | 0.0 | 0.0 | 2672 | 7.2250 | C |
| 604 | 3.0 | Abelseth, Miss. Karen Marie | female | 16.0 | 0.0 | 0.0 | 348125 | 7.6500 | S |
Scikit-learn deals with learning information from one or more datasets that are represented as 2D arrays. They can be understood as a list of multi-dimensional observations. We say that the first axis of these arrays is the samples axis, while the second is the features axis. > (n_samples, n_features)
An estimator is any object that learns from data; it may be a classification, regression or clustering algorithm or a transformer that extracts/filters useful features from raw data.
All estimator objects expose a fit method that takes a dataset (usually a 2-d array) > estimator.fit(data)
An estimator supporting transform and/or fit_transform methods.
A transformer, transforms the input, usually only X, into some transformed space. Output is an array or sparse matrix of length n_samples and with the number of columns fixed after fitting.
The fit method is provided on every estimator. It usually takes some samples X, targets y if the model is supervised, and potentially other sample properties such as sample_weight.
It should: * clear any prior attributes stored on the estimator, unless warm_start is used * validate and interpret any parameters, ideally raising an error if invalid * validate the input data * estimate and store model attributes from the estimated parameters and provided data; and * return the now fitted estimator to facilitate method chaining
Note: * Fitting = Calling fit (or fit_transform, fit_predict) method on an estimator. * Fitted = The state of an estimator after fitting.
class sklearn.pipeline.Pipeline(steps, *, memory=None, verbose=False)
It is a pipeline of transformers with a final estimator.
It sequentially applies a list of transforms and a final estimator. Intermediate steps of the pipeline must be transforms, that is, they must implement fit and transform methods. The final estimator only needs to implement fit.
Lets create a simple pipeline to better understand its componets. Steps in our pipeline will be * replace missing values using the mean along each numerical feature column; and * then scale them
from sklearn.pipeline import Pipeline
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import StandardScaler
pipe = Pipeline(steps=[
('imputer', SimpleImputer()),
('scaler', StandardScaler())
])
# our first pipeline has been initialized
pipePipeline(steps=[('imputer', SimpleImputer()), ('scaler', StandardScaler())])We can also visualize the pipeline as a diagram. It has two steps: imputer and scaler in sequence.
Pipeline(steps=[('imputer', SimpleImputer()), ('scaler', StandardScaler())])Please rerun this cell to show the HTML repr or trust the notebook.Pipeline(steps=[('imputer', SimpleImputer()), ('scaler', StandardScaler())])SimpleImputer()
StandardScaler()
now lets call fit_transform method to run this pipeline, and preprocess our loaded data
--------------------------------------------------------------------------- ValueError Traceback (most recent call last) Input In [8], in <module> 1 #collapse-output ----> 2 pipe.fit_transform(X_train, y_train) File ~\anaconda3\envs\sc_mlflow\lib\site-packages\sklearn\pipeline.py:426, in Pipeline.fit_transform(self, X, y, **fit_params) 399 """Fit the model and transform with the final estimator. 400 401 Fits all the transformers one after the other and transform the (...) 423 Transformed samples. 424 """ 425 fit_params_steps = self._check_fit_params(**fit_params) --> 426 Xt = self._fit(X, y, **fit_params_steps) 428 last_step = self._final_estimator 429 with _print_elapsed_time("Pipeline", self._log_message(len(self.steps) - 1)): File ~\anaconda3\envs\sc_mlflow\lib\site-packages\sklearn\pipeline.py:348, in Pipeline._fit(self, X, y, **fit_params_steps) 346 cloned_transformer = clone(transformer) 347 # Fit or load from cache the current transformer --> 348 X, fitted_transformer = fit_transform_one_cached( 349 cloned_transformer, 350 X, 351 y, 352 None, 353 message_clsname="Pipeline", 354 message=self._log_message(step_idx), 355 **fit_params_steps[name], 356 ) 357 # Replace the transformer of the step with the fitted 358 # transformer. This is necessary when loading the transformer 359 # from the cache. 360 self.steps[step_idx] = (name, fitted_transformer) File ~\anaconda3\envs\sc_mlflow\lib\site-packages\joblib\memory.py:352, in NotMemorizedFunc.__call__(self, *args, **kwargs) 351 def __call__(self, *args, **kwargs): --> 352 return self.func(*args, **kwargs) File ~\anaconda3\envs\sc_mlflow\lib\site-packages\sklearn\pipeline.py:893, in _fit_transform_one(transformer, X, y, weight, message_clsname, message, **fit_params) 891 with _print_elapsed_time(message_clsname, message): 892 if hasattr(transformer, "fit_transform"): --> 893 res = transformer.fit_transform(X, y, **fit_params) 894 else: 895 res = transformer.fit(X, y, **fit_params).transform(X) File ~\anaconda3\envs\sc_mlflow\lib\site-packages\sklearn\base.py:855, in TransformerMixin.fit_transform(self, X, y, **fit_params) 852 return self.fit(X, **fit_params).transform(X) 853 else: 854 # fit method of arity 2 (supervised transformation) --> 855 return self.fit(X, y, **fit_params).transform(X) File ~\anaconda3\envs\sc_mlflow\lib\site-packages\sklearn\impute\_base.py:319, in SimpleImputer.fit(self, X, y) 302 def fit(self, X, y=None): 303 """Fit the imputer on `X`. 304 305 Parameters (...) 317 Fitted estimator. 318 """ --> 319 X = self._validate_input(X, in_fit=True) 321 # default fill_value is 0 for numerical input and "missing_value" 322 # otherwise 323 if self.fill_value is None: File ~\anaconda3\envs\sc_mlflow\lib\site-packages\sklearn\impute\_base.py:285, in SimpleImputer._validate_input(self, X, in_fit) 279 if "could not convert" in str(ve): 280 new_ve = ValueError( 281 "Cannot use {} strategy with non-numeric data:\n{}".format( 282 self.strategy, ve 283 ) 284 ) --> 285 raise new_ve from None 286 else: 287 raise ve ValueError: Cannot use mean strategy with non-numeric data: could not convert string to float: "McCarthy, Miss. Catherine 'Katie'"
Aaargh! this is not what we intended. Let us try to understand why our pipeline did not work and then fix it. The exception message says:
ValueError: Cannot use mean strategy with non-numeric data: could not convert string to float: "McCarthy, Miss. Catherine 'Katie'From the error message we can deduce that Pipeline is trying to apply its transformers on all columns in the dataset. This was not our intention, as we wanted to apply the transformers to numeric data only. Let’s limit our simple pipeline to numerical columns and run again.
array([[ 0. , -0.49963779],
[-0.43641134, -0.09097855],
[-1.44872891, -0.01824953],
...,
[-0.98150542, -0.49349894],
[-0.82576425, -0.44336498],
[-0.59215251, -0.49349894]])Alright, our pipeline has run now and we can also observe a few outcomes. * When we apply a pipeline to a dataset it will run transformers to all features in the dataset. * Output from one transformer will be passed on to the next one until we reach the end of the pipeline * If we want to apply different transformers for numerical and categorical features (heterogeneous data) then the pipeline will not work for us. We would have to create separate pipelines for the different feature sets and then join the output.
To overcome the limitation of a pipeline for heterogeneous data, Sklearn recommends using ColumnTransformer. With ColumnTransformer we can provide column names against the transformers on which we want to apply them.
Let’s see our first ColumnTransformer in action.
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import OneHotEncoder
# Note the sequence when creating a ColumnTransformer
# 1. a name for the transformer
# 2. the transformer
# 3. the column names
pipe = ColumnTransformer([
('standardscaler', StandardScaler(), ['age', 'fare'] ),
('onehotencoder', OneHotEncoder(), ['sex'])
])
pipeColumnTransformer(transformers=[('standardscaler', StandardScaler(),
['age', 'fare']),
('onehotencoder', OneHotEncoder(), ['sex'])])Please rerun this cell to show the HTML repr or trust the notebook.ColumnTransformer(transformers=[('standardscaler', StandardScaler(),
['age', 'fare']),
('onehotencoder', OneHotEncoder(), ['sex'])])['age', 'fare']
StandardScaler()
['sex']
OneHotEncoder()
array([[ nan, -0.49939913, 1. , 0. ],
[-0.39043136, -0.09093509, 1. , 0. ],
[-1.2960919 , -0.01824081, 1. , 0. ],
...,
[-0.87809473, -0.49326321, 0. , 1. ],
[-0.73876234, -0.4431532 , 0. , 1. ],
[-0.52976375, -0.49326321, 0. , 1. ]])At this point I will also introduce a very useful function get_feature_names_out(input_features=None). Using this method we can get output feature names as well.
array(['standardscaler__age', 'standardscaler__fare',
'onehotencoder__sex_female', 'onehotencoder__sex_male'],
dtype=object)Notice the output * Output feature names appear as <transformer_name>__<feature_name> * For OneHotEncoded feature “sex”, output feature names have the label attached to them
Sklean also provides a wrapper function for ColumnTransformer where we don’t have to provide names for the transformers.
from sklearn.compose import make_column_transformer
# Note the sequence when using make_column_transformer
# 1. the transformer
# 2. the column names
pipe = make_column_transformer(
(StandardScaler(), ['age', 'fare']),
(OneHotEncoder(), ['sex'] ),
verbose_feature_names_out=False # to keep output feature names simple
)
pipeColumnTransformer(transformers=[('standardscaler', StandardScaler(),
['age', 'fare']),
('onehotencoder', OneHotEncoder(), ['sex'])],
verbose_feature_names_out=False)Please rerun this cell to show the HTML repr or trust the notebook.ColumnTransformer(transformers=[('standardscaler', StandardScaler(),
['age', 'fare']),
('onehotencoder', OneHotEncoder(), ['sex'])],
verbose_feature_names_out=False)['age', 'fare']
StandardScaler()
['sex']
OneHotEncoder()
array([[ nan, -0.49939913, 1. , 0. ],
[-0.39043136, -0.09093509, 1. , 0. ],
[-1.2960919 , -0.01824081, 1. , 0. ],
...,
[-0.87809473, -0.49326321, 0. , 1. ],
[-0.73876234, -0.4431532 , 0. , 1. ],
[-0.52976375, -0.49326321, 0. , 1. ]])So our ColumnTransformer is working. But we have a few more questions to address. * Why is the output from our pipeline or ColumnTransformer not shown as a dataframe with output features nicely separated in different columns? * Our input dataset had more features besides age, fare, and sex. Why are they not present in the output? * What happens if I change the sequence of transformers, and feature names in my ColumnTransformer?
In the coming sections, we will try to address these questions.
The output from a pipeline or a ColumnTransformer is an nd-array where the first index is the number of samples, and second index are the output features (n_samples, n_output_features). Since we are only getting numpy array as an output, we are losing information about the column names.
We have already seen that we can get the output feature names using method get_feature_names_out. But this time let’s try to analyze our ColumnsTransformer more closely. The transformer attributes discussed here also applies to Pipeline object.
ColumnTransformer(transformers=[('standardscaler', StandardScaler(),
['age', 'fare']),
('onehotencoder', OneHotEncoder(), ['sex'])],
verbose_feature_names_out=False)ColumnTransformer has an attribute ‘transformers’ that is keeping a list of all the provided transformers. Let’s print it.
[('standardscaler', StandardScaler(), ['age', 'fare']),
('onehotencoder', OneHotEncoder(), ['sex'])]These are the transformers list at the initialization time. If we want to check the transformers after fit function has been called, then we need to print a different attribute transformers_.
[('standardscaler', StandardScaler(), ['age', 'fare']),
('onehotencoder', OneHotEncoder(), ['sex']),
('remainder', 'drop', [0, 1, 4, 5, 6, 8])]You can see the difference. There is an extra transformer with the name remainder at the end. It was not present at the initialization time. What it does is that it drops all remaining columns from the dataset that have not been explicitly used in the ColumnTransformer. Since, at the initialization time, ColumnTransformer does not know about the other columns that it needs to drop this transformer is missing. During fit it sees the dataset and knows about the other columns, it then keeps a list of them to drop (0, 1, 4, 5, 6, 8).
We can also index through the transformers as well to fetch anyone from the list.
('onehotencoder', OneHotEncoder(), ['sex'])Notice the tuple sequence. * First is the name * Second is the transformer * Third are the column names
We can also call get_feature_names_out method on a separate transformer from the list.
array(['sex_female', 'sex_male'], dtype=object)##
# output features from last tranformer
pipe.transformers_[-1][1].get_feature_names_out()
# No. We cannot do this on last transformer (remainder).--------------------------------------------------------------------------- AttributeError Traceback (most recent call last) Input In [22], in <module> 1 #collapse-output 2 # output features from last tranformer ----> 3 pipe.transformers_[-1][1].get_feature_names_out() AttributeError: 'str' object has no attribute 'get_feature_names_out'
We now have output feature names, and the output (nd-array). Can we convert them to a DataFrame?
import pandas as pd
temp = pipe.fit_transform(X_train, y_train)
col_names = pipe.get_feature_names_out()
output = pd.DataFrame(temp.T, col_names).T
output.head()| age | fare | sex_female | sex_male | |
|---|---|---|---|---|
| 0 | NaN | -0.499399 | 1.0 | 0.0 |
| 1 | -0.390431 | -0.090935 | 1.0 | 0.0 |
| 2 | -1.296092 | -0.018241 | 1.0 | 0.0 |
| 3 | -0.320765 | -0.510137 | 0.0 | 1.0 |
| 4 | -0.947761 | -0.501444 | 1.0 | 0.0 |
We know how to convert the transformer output to a DataFrame. It would be much simpler if we don’t have to do an extra step, and can directly get a Dataframe from our fitted ColumnTransformer.
For this we can take the help of FunctionTransformer > A FunctionTransformer forwards its X (and optionally y) arguments to a user-defined function or function object and returns the result of this function.
Let’s see a FunctionTransformer in action.
from sklearn.preprocessing import FunctionTransformer
preprocessor = make_column_transformer(
(StandardScaler(), ['age', 'fare']),
(OneHotEncoder(), ['sex'] ),
verbose_feature_names_out=False
)
pipe = Pipeline([
("preprocess", preprocessor),
("dataframer",FunctionTransformer(lambda x: pd.DataFrame(x, columns = preprocessor.get_feature_names_out())))
])
set_config(display="diagram")
pipePipeline(steps=[('preprocess',
ColumnTransformer(transformers=[('standardscaler',
StandardScaler(),
['age', 'fare']),
('onehotencoder',
OneHotEncoder(), ['sex'])],
verbose_feature_names_out=False)),
('dataframer',
FunctionTransformer(func=<function <lambda> at 0x0000016196A1B040>))])Please rerun this cell to show the HTML repr or trust the notebook.Pipeline(steps=[('preprocess',
ColumnTransformer(transformers=[('standardscaler',
StandardScaler(),
['age', 'fare']),
('onehotencoder',
OneHotEncoder(), ['sex'])],
verbose_feature_names_out=False)),
('dataframer',
FunctionTransformer(func=<function <lambda> at 0x0000016196A1B040>))])ColumnTransformer(transformers=[('standardscaler', StandardScaler(),
['age', 'fare']),
('onehotencoder', OneHotEncoder(), ['sex'])],
verbose_feature_names_out=False)['age', 'fare']
StandardScaler()
['sex']
OneHotEncoder()
FunctionTransformer(func=<function <lambda> at 0x0000016196A1B040>)
Notice that we have applied FunctionTransformer after ColumnTransformer in a Pipeline. When we fit our pipeline on the dataset, ColumnTransformer will be fitted first and then the FunctionTransformer. Since the ColumnTransformer has been fitted first, we will be able to call get_feature_names_out on it while passing data to FunctionTransformer.
| age | fare | sex_female | sex_male | |
|---|---|---|---|---|
| 0 | NaN | -0.499399 | 1.0 | 0.0 |
| 1 | -0.390431 | -0.090935 | 1.0 | 0.0 |
| 2 | -1.296092 | -0.018241 | 1.0 | 0.0 |
| 3 | -0.320765 | -0.510137 | 0.0 | 1.0 |
| 4 | -0.947761 | -0.501444 | 1.0 | 0.0 |
This is looking good. We are now getting back a dataframe directly from the pipeline. With a dataframe it is a lot easier to view and verify the output from the preprocessor.
But we have to be very careful with FunctionTransformer. In Sklearn docs, it says
Note: If a lambda is used as the function, then the resulting transformer will not be pickleable.
Huh! that is a very concerning point. We have also used a lambda function, and we will not be able to pickle it. Let’s check it first.
--------------------------------------------------------------------------- PicklingError Traceback (most recent call last) Input In [26], in <module> 1 import pickle 3 # save our pipeline ----> 4 s1 = pickle.dumps(pipe) 6 # reload it 7 s2 = pickle.loads(s1) PicklingError: Can't pickle <function <lambda> at 0x0000016196A1B040>: attribute lookup <lambda> on __main__ failed
The documentation was right about it. We have used a Lambda function in our FunctionTranformer and we got a pickle error. Since, the limitation is said for Lambda function, changing it with a normal function should work. Let’s do that.
def get_dataframe(X, transformer):
"""
x: an nd-array
transformer: fitted transformer
"""
col_names = transformer.get_feature_names_out()
output = pd.DataFrame(X.T, col_names).T
return output
preprocessor = make_column_transformer(
(StandardScaler(), ['age', 'fare']),
(OneHotEncoder(), ['sex'] ),
verbose_feature_names_out=False
)
dataframer = FunctionTransformer(func=get_dataframe, kw_args={"transformer": preprocessor})
pipe = Pipeline([
("preprocess", preprocessor),
("dataframer", dataframer)
])
set_config(display="diagram")
pipePipeline(steps=[('preprocess',
ColumnTransformer(transformers=[('standardscaler',
StandardScaler(),
['age', 'fare']),
('onehotencoder',
OneHotEncoder(), ['sex'])],
verbose_feature_names_out=False)),
('dataframer',
FunctionTransformer(func=<function get_dataframe at 0x00000161953C3670>,
kw_args={'transformer': ColumnTransformer(transformers=[('standardscaler',
StandardScaler(),
['age',
'fare']),
('onehotencoder',
OneHotEncoder(),
['sex'])],
verbose_feature_names_out=False)}))])Please rerun this cell to show the HTML repr or trust the notebook.Pipeline(steps=[('preprocess',
ColumnTransformer(transformers=[('standardscaler',
StandardScaler(),
['age', 'fare']),
('onehotencoder',
OneHotEncoder(), ['sex'])],
verbose_feature_names_out=False)),
('dataframer',
FunctionTransformer(func=<function get_dataframe at 0x00000161953C3670>,
kw_args={'transformer': ColumnTransformer(transformers=[('standardscaler',
StandardScaler(),
['age',
'fare']),
('onehotencoder',
OneHotEncoder(),
['sex'])],
verbose_feature_names_out=False)}))])ColumnTransformer(transformers=[('standardscaler', StandardScaler(),
['age', 'fare']),
('onehotencoder', OneHotEncoder(), ['sex'])],
verbose_feature_names_out=False)['age', 'fare']
StandardScaler()
['sex']
OneHotEncoder()
FunctionTransformer(func=<function get_dataframe at 0x00000161953C3670>,
kw_args={'transformer': ColumnTransformer(transformers=[('standardscaler',
StandardScaler(),
['age',
'fare']),
('onehotencoder',
OneHotEncoder(),
['sex'])],
verbose_feature_names_out=False)})Notice the arguments for FunctionTransformer in the above code. * first argument is the function to be called * second argument are the parameters to be passed to our function
The sequence of arguments for the callable function will be * first argument will be the output from any previous step in the pipeline (if there is any). In our case, it is nd-array coming from ColumnTransformer. It will be mapped to X. We don’t have to do anything about it. * second argument (if any) we want to pass to function. In our case we need it to be the fitted transformer from the previous step so we have explicitly passed it using kw_args as key-value pair. Where key name is the same as callable method argument name (‘transformer’ in our case).
Now let’s do our pickle test one more time.
Pipeline(steps=[('preprocess',
ColumnTransformer(transformers=[('standardscaler',
StandardScaler(),
['age', 'fare']),
('onehotencoder',
OneHotEncoder(), ['sex'])],
verbose_feature_names_out=False)),
('dataframer',
FunctionTransformer(func=<function get_dataframe at 0x00000161953C3670>,
kw_args={'transformer': ColumnTransformer(transformers=[('standardscaler',
StandardScaler(),
['age',
'fare']),
('onehotencoder',
OneHotEncoder(),
['sex'])],
verbose_feature_names_out=False)}))])Please rerun this cell to show the HTML repr or trust the notebook.Pipeline(steps=[('preprocess',
ColumnTransformer(transformers=[('standardscaler',
StandardScaler(),
['age', 'fare']),
('onehotencoder',
OneHotEncoder(), ['sex'])],
verbose_feature_names_out=False)),
('dataframer',
FunctionTransformer(func=<function get_dataframe at 0x00000161953C3670>,
kw_args={'transformer': ColumnTransformer(transformers=[('standardscaler',
StandardScaler(),
['age',
'fare']),
('onehotencoder',
OneHotEncoder(),
['sex'])],
verbose_feature_names_out=False)}))])ColumnTransformer(transformers=[('standardscaler', StandardScaler(),
['age', 'fare']),
('onehotencoder', OneHotEncoder(), ['sex'])],
verbose_feature_names_out=False)['age', 'fare']
StandardScaler()
['sex']
OneHotEncoder()
FunctionTransformer(func=<function get_dataframe at 0x00000161953C3670>,
kw_args={'transformer': ColumnTransformer(transformers=[('standardscaler',
StandardScaler(),
['age',
'fare']),
('onehotencoder',
OneHotEncoder(),
['sex'])],
verbose_feature_names_out=False)})Alright, no more issues so let’s proceed to our next question.
By default, only the specified columns in transformers are transformed and combined in the output, and the non-specified columns are dropped. (default of remainder='drop'). By specifying remainder='passthrough', all remaining columns that were not specified in transformers will be automatically passed through. This subset of columns is concatenated with the output of the transformers.
Let’s see it in action.
preprocessor = make_column_transformer(
(StandardScaler(), ['age', 'fare']),
(OneHotEncoder(), ['sex'] ),
verbose_feature_names_out=False,
remainder='passthrough'
)
# get_dataframe is already defined in last section. Intentionally omitted here.
dataframer = FunctionTransformer(func=get_dataframe, kw_args={"transformer": preprocessor})
pipe = Pipeline([
("preprocess", preprocessor),
("dataframer", dataframer)
])
temp = pipe.fit_transform(X_train, y_train)
temp.head()| age | fare | sex_female | sex_male | pclass | name | sibsp | parch | ticket | embarked | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | NaN | -0.499399 | 1.0 | 0.0 | 3.0 | McCarthy, Miss. Catherine 'Katie' | 0.0 | 0.0 | 383123 | Q |
| 1 | -0.390431 | -0.090935 | 1.0 | 0.0 | 2.0 | del Carlo, Mrs. Sebastiano (Argenia Genovesi) | 1.0 | 0.0 | SC/PARIS 2167 | C |
| 2 | -1.296092 | -0.018241 | 1.0 | 0.0 | 3.0 | Andersson, Miss. Sigrid Elisabeth | 4.0 | 2.0 | 347082 | S |
| 3 | -0.320765 | -0.510137 | 0.0 | 1.0 | 3.0 | Saad, Mr. Khalil | 0.0 | 0.0 | 2672 | C |
| 4 | -0.947761 | -0.501444 | 1.0 | 0.0 | 3.0 | Abelseth, Miss. Karen Marie | 0.0 | 0.0 | 348125 | S |
We have our remaining features back now, so let’s proceed to our next question.
It is better to make some changes and then see the results. I am making two changes in ColumnTransformer 1. Changed the order of transformers (OHE before scaling) 2. Changed the order of features inside the transformer (‘fare’ before ‘age’)
preprocessor = make_column_transformer(
(OneHotEncoder(), ['sex'] ),
(StandardScaler(), ['fare', 'age']),
verbose_feature_names_out=False,
remainder='passthrough'
)
# get_dataframe is already defined in last section. Intentionally omitted here.
dataframer = FunctionTransformer(func=get_dataframe, kw_args={"transformer": preprocessor})
pipe = Pipeline([
("preprocess", preprocessor),
("dataframer", dataframer)
])
temp = pipe.fit_transform(X_train, y_train)
temp.head()| sex_female | sex_male | fare | age | pclass | name | sibsp | parch | ticket | embarked | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1.0 | 0.0 | -0.499399 | NaN | 3.0 | McCarthy, Miss. Catherine 'Katie' | 0.0 | 0.0 | 383123 | Q |
| 1 | 1.0 | 0.0 | -0.090935 | -0.390431 | 2.0 | del Carlo, Mrs. Sebastiano (Argenia Genovesi) | 1.0 | 0.0 | SC/PARIS 2167 | C |
| 2 | 1.0 | 0.0 | -0.018241 | -1.296092 | 3.0 | Andersson, Miss. Sigrid Elisabeth | 4.0 | 2.0 | 347082 | S |
| 3 | 0.0 | 1.0 | -0.510137 | -0.320765 | 3.0 | Saad, Mr. Khalil | 0.0 | 0.0 | 2672 | C |
| 4 | 1.0 | 0.0 | -0.501444 | -0.947761 | 3.0 | Abelseth, Miss. Karen Marie | 0.0 | 0.0 | 348125 | S |
We can see that changing the sequence in ColumnTransformer does change the output. Also note * Specified columns in transformers are transformed and combined in the output * Transformers sequence in ColumnTransformer also represents the columns sequence in the output * When remainder=passthrough is used then remaining columns will be appended at the end. Remainder columns sequence will be same as in the input.
Let’s assume we have more requirements this time. I want * for numerical features (age, fare): impute the missing values first, and then scale them * for categorical features (sex): one hot encode them
Our pipeline will look like this.
#collapse-output
numeric_features = ["age", "fare"]
numeric_transformer = Pipeline(
steps=[
("imputer", SimpleImputer(strategy="median")),
("scaler", StandardScaler())
])
categorical_features = ["sex"]
categorical_transformer = OneHotEncoder(handle_unknown="ignore")
preprocessor = make_column_transformer(
(numeric_transformer, numeric_features), # note "numeric_transformer" is a pipeline this time
(categorical_transformer, categorical_features),
remainder='passthrough',
verbose_feature_names_out=False
)
dataframer = FunctionTransformer(func=get_dataframe, kw_args={"transformer": preprocessor})
pipe = Pipeline([
("preprocess", preprocessor),
("dataframer", dataframer)
])
temp = pipe.fit_transform(X_train, y_train)
temp.head()--------------------------------------------------------------------------- AttributeError Traceback (most recent call last) Input In [31], in <module> 18 dataframer = FunctionTransformer(func=get_dataframe, kw_args={"transformer": preprocessor}) 19 pipe = Pipeline([ 20 ("preprocess", preprocessor), 21 ("dataframer", dataframer) 22 ]) ---> 24 temp = pipe.fit_transform(X_train, y_train) 25 temp.head() File ~\anaconda3\envs\sc_mlflow\lib\site-packages\sklearn\pipeline.py:434, in Pipeline.fit_transform(self, X, y, **fit_params) 432 fit_params_last_step = fit_params_steps[self.steps[-1][0]] 433 if hasattr(last_step, "fit_transform"): --> 434 return last_step.fit_transform(Xt, y, **fit_params_last_step) 435 else: 436 return last_step.fit(Xt, y, **fit_params_last_step).transform(Xt) File ~\anaconda3\envs\sc_mlflow\lib\site-packages\sklearn\base.py:855, in TransformerMixin.fit_transform(self, X, y, **fit_params) 852 return self.fit(X, **fit_params).transform(X) 853 else: 854 # fit method of arity 2 (supervised transformation) --> 855 return self.fit(X, y, **fit_params).transform(X) File ~\anaconda3\envs\sc_mlflow\lib\site-packages\sklearn\preprocessing\_function_transformer.py:182, in FunctionTransformer.transform(self, X) 169 """Transform X using the forward function. 170 171 Parameters (...) 179 Transformed input. 180 """ 181 X = self._check_input(X, reset=False) --> 182 return self._transform(X, func=self.func, kw_args=self.kw_args) File ~\anaconda3\envs\sc_mlflow\lib\site-packages\sklearn\preprocessing\_function_transformer.py:205, in FunctionTransformer._transform(self, X, func, kw_args) 202 if func is None: 203 func = _identity --> 205 return func(X, **(kw_args if kw_args else {})) Input In [27], in get_dataframe(X, transformer) 1 def get_dataframe(X, transformer): 2 """ 3 x: an nd-array 4 transformer: fitted transformer 5 """ ----> 6 col_names = transformer.get_feature_names_out() 7 output = pd.DataFrame(X.T, col_names).T 8 return output File ~\anaconda3\envs\sc_mlflow\lib\site-packages\sklearn\compose\_column_transformer.py:481, in ColumnTransformer.get_feature_names_out(self, input_features) 479 transformer_with_feature_names_out = [] 480 for name, trans, column, _ in self._iter(fitted=True): --> 481 feature_names_out = self._get_feature_name_out_for_transformer( 482 name, trans, column, input_features 483 ) 484 if feature_names_out is None: 485 continue File ~\anaconda3\envs\sc_mlflow\lib\site-packages\sklearn\compose\_column_transformer.py:454, in ColumnTransformer._get_feature_name_out_for_transformer(self, name, trans, column, feature_names_in) 450 if isinstance(column, Iterable) and not all( 451 isinstance(col, str) for col in column 452 ): 453 column = _safe_indexing(feature_names_in, column) --> 454 return trans.get_feature_names_out(column) File ~\anaconda3\envs\sc_mlflow\lib\site-packages\sklearn\pipeline.py:751, in Pipeline.get_feature_names_out(self, input_features) 749 for _, name, transform in self._iter(): 750 if not hasattr(transform, "get_feature_names_out"): --> 751 raise AttributeError( 752 "Estimator {} does not provide get_feature_names_out. " 753 "Did you mean to call pipeline[:-1].get_feature_names_out" 754 "()?".format(name) 755 ) 756 feature_names_out = transform.get_feature_names_out(feature_names_out) 757 return feature_names_out AttributeError: Estimator imputer does not provide get_feature_names_out. Did you mean to call pipeline[:-1].get_feature_names_out()?
Oh geez! What went wrong this time. The error message says
AttributeError: Estimator imputer does not provide get_feature_names_out. Did you mean to call pipeline[:-1].get_feature_names_out()?From the error message I am getting that > Estimator imputer does not provide get_feature_names_out
Hmmm, this is strange. Why is this estimator missing a very useful function? Let’s check the docs first on SimpleImputer. For the docs I indeed could not find this method get_feature_names_out() for this transformer. A little googling lead me to this Sklearn Github issue page Implement get_feature_names_out for all estimators. Developers are actively adding get_feature_names_out() to all estimators and transformers, and it looks like this feature has not been implemented for SimpleImputer till Sklearn version==1.0.2.
But no worries we can overcome this limitation, and implement this feature ourselves through a custom transformer.
We can create a custom transformer or an estimator simply by inheriting a class from BaseEstimator and optionally the mixin classes in sklearn.base. Sklean provides a template that we can use to create our custom transformer. Template link is here: https://github.com/scikit-learn-contrib/project-template/blob/master/skltemplate/_template.py#L146
Let us use the same pipeline as in last cell but replace SimpleImputer with a custom one.
from sklearn.base import BaseEstimator, TransformerMixin
class SimpleImputerCustom(BaseEstimator, TransformerMixin):
def __init__(self, strategy='mean'):
self.strategy = strategy
self.imputer = SimpleImputer(strategy=self.strategy)
def fit(self, X, y):
self.imputer.fit(X, y)
return self
def transform(self, X):
return self.imputer.transform(X)
def get_feature_names_out(self, input_features=None):
# we have returned the input features name as out features will have the same name
return input_features
numeric_features = ["age", "fare"]
numeric_transformer = Pipeline(
steps=[
("imputer", SimpleImputerCustom(strategy='mean')),
("scaler", StandardScaler())
]
)
categorical_features = ["sex"]
categorical_transformer = OneHotEncoder(handle_unknown="ignore")
preprocessor = make_column_transformer(
(numeric_transformer, numeric_features), # note "numeric_transformer" is a pipeline
(categorical_transformer, categorical_features),
remainder='passthrough',
verbose_feature_names_out=False
)
dataframer = FunctionTransformer(func=get_dataframe, kw_args={"transformer": preprocessor})
pipe = Pipeline([
("preprocess", preprocessor),
("dataframer", dataframer)
])
temp = pipe.fit_transform(X_train, y_train)
temp.head()| age | fare | sex_female | sex_male | pclass | name | sibsp | parch | ticket | embarked | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0 | -0.499638 | 1.0 | 0.0 | 3.0 | McCarthy, Miss. Catherine 'Katie' | 0.0 | 0.0 | 383123 | Q |
| 1 | -0.436411 | -0.090979 | 1.0 | 0.0 | 2.0 | del Carlo, Mrs. Sebastiano (Argenia Genovesi) | 1.0 | 0.0 | SC/PARIS 2167 | C |
| 2 | -1.448729 | -0.01825 | 1.0 | 0.0 | 3.0 | Andersson, Miss. Sigrid Elisabeth | 4.0 | 2.0 | 347082 | S |
| 3 | -0.358541 | -0.510381 | 0.0 | 1.0 | 3.0 | Saad, Mr. Khalil | 0.0 | 0.0 | 2672 | C |
| 4 | -1.059376 | -0.501684 | 1.0 | 0.0 | 3.0 | Abelseth, Miss. Karen Marie | 0.0 | 0.0 | 348125 | S |
So far, so good! Let’s assume that we have another requirement and it is about feature engineering. We have to combine ‘sibsp’ and ‘parch’ into two new features: family_size and is_alone.
Let’s implement this now.
class FamilyFeatureTransformer(BaseEstimator, TransformerMixin):
def __init__(self):
pass
def fit(self, X, y):
return self
def transform(self, X):
X['family_size'] = X['parch'] + X['sibsp']
X.drop(['parch', 'sibsp'], axis=1, inplace=True) # we can drop this feature now
X['is_alone'] = 1
X.loc[X['family_size'] > 1, 'is_alone'] = 0
return X
def get_feature_names_out(self, input_features=None):
# this time we have created new features. Their names are different from input features.
# so we have explicitly mentioned them here.
return ['family_size', 'is_alone']
numeric_features = ["age", "fare"]
numeric_transformer = Pipeline(
steps=[
("imputer", SimpleImputerCustom(strategy='mean')),
("scaler", StandardScaler())
]
)
categorical_features = ["sex"]
categorical_transformer = OneHotEncoder(handle_unknown="ignore")
family_features = ["parch", "sibsp"]
family_transformer = FamilyFeatureTransformer()
preprocessor = make_column_transformer(
(numeric_transformer, numeric_features),
(categorical_transformer, categorical_features),
(family_transformer, family_features),
remainder='drop', # let's drop extra features this time
verbose_feature_names_out=False
)
dataframer = FunctionTransformer(func=get_dataframe, kw_args={"transformer": preprocessor})
pipe = Pipeline([
("preprocess", preprocessor),
("dataframer", dataframer)
])
temp = pipe.fit_transform(X_train, y_train)
temp.head()| age | fare | sex_female | sex_male | family_size | is_alone | |
|---|---|---|---|---|---|---|
| 0 | 0.000000 | -0.499638 | 1.0 | 0.0 | 0.0 | 1.0 |
| 1 | -0.436411 | -0.090979 | 1.0 | 0.0 | 1.0 | 1.0 |
| 2 | -1.448729 | -0.018250 | 1.0 | 0.0 | 6.0 | 0.0 |
| 3 | -0.358541 | -0.510381 | 0.0 | 1.0 | 0.0 | 1.0 |
| 4 | -1.059376 | -0.501684 | 1.0 | 0.0 | 0.0 | 1.0 |
Alright, we have our required features ready and we can now pass them to a classifier. Let’s use RandomForrest as our classifier and run our pipeline with it.
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score
# 'preprocessor' and 'dataframer' are already declared in last section, and intentionally omitted here.
pipe = Pipeline([
("preprocess", preprocessor),
("dataframer", dataframer),
('rf_estimator', RandomForestClassifier())
])
temp = pipe.fit_transform(X_train, y_train)
temp.head()--------------------------------------------------------------------------- AttributeError Traceback (most recent call last) Input In [34], in <module> 4 # 'preprocessor' and 'dataframer' are already declared in last section, and intentionally omitted here. 5 pipe = Pipeline([ 6 ("preprocess", preprocessor), 7 ("dataframer", dataframer), 8 ('rf_estimator', RandomForestClassifier()) 9 10 ]) ---> 12 temp = pipe.fit_transform(X_train, y_train) 13 temp.head() File ~\anaconda3\envs\sc_mlflow\lib\site-packages\sklearn\pipeline.py:436, in Pipeline.fit_transform(self, X, y, **fit_params) 434 return last_step.fit_transform(Xt, y, **fit_params_last_step) 435 else: --> 436 return last_step.fit(Xt, y, **fit_params_last_step).transform(Xt) AttributeError: 'RandomForestClassifier' object has no attribute 'transform'
Okay, looks like we have made a mistake here. Error message is saying
AttributeError: 'RandomForestClassifier' object has no attribute 'transform'I get that. In our pipeline we have an estimator that does not have a transform method defined for it. We should use predict method instead.
Note: * Estimators implement predict method (Template reference Estimator, Template reference Classifier) * Transformers implement transform method (Template reference Transformer) * fit_transform is same calling fit and then transform
Let us fix the error and run our pipeline again.
##
# pipeline created in last section and intentionally omitted here.
pipe.fit(X_train, y_train)
y_pred = pipe.predict(X_test)
accuracy_score(y_test, y_pred)0.7862595419847328Let’s see how our final pipeline looks visually.
Pipeline(steps=[('preprocess',
ColumnTransformer(transformers=[('pipeline',
Pipeline(steps=[('imputer',
SimpleImputerCustom()),
('scaler',
StandardScaler())]),
['age', 'fare']),
('onehotencoder',
OneHotEncoder(handle_unknown='ignore'),
['sex']),
('familyfeaturetransformer',
FamilyFeatureTransformer(),
['parch', 'sibsp'])],
verbose_feature_names_out=False)),
('data...
kw_args={'transformer': ColumnTransformer(transformers=[('pipeline',
Pipeline(steps=[('imputer',
SimpleImputerCustom()),
('scaler',
StandardScaler())]),
['age',
'fare']),
('onehotencoder',
OneHotEncoder(handle_unknown='ignore'),
['sex']),
('familyfeaturetransformer',
FamilyFeatureTransformer(),
['parch',
'sibsp'])],
verbose_feature_names_out=False)})),
('rf_estimator', RandomForestClassifier())])Please rerun this cell to show the HTML repr or trust the notebook.Pipeline(steps=[('preprocess',
ColumnTransformer(transformers=[('pipeline',
Pipeline(steps=[('imputer',
SimpleImputerCustom()),
('scaler',
StandardScaler())]),
['age', 'fare']),
('onehotencoder',
OneHotEncoder(handle_unknown='ignore'),
['sex']),
('familyfeaturetransformer',
FamilyFeatureTransformer(),
['parch', 'sibsp'])],
verbose_feature_names_out=False)),
('data...
kw_args={'transformer': ColumnTransformer(transformers=[('pipeline',
Pipeline(steps=[('imputer',
SimpleImputerCustom()),
('scaler',
StandardScaler())]),
['age',
'fare']),
('onehotencoder',
OneHotEncoder(handle_unknown='ignore'),
['sex']),
('familyfeaturetransformer',
FamilyFeatureTransformer(),
['parch',
'sibsp'])],
verbose_feature_names_out=False)})),
('rf_estimator', RandomForestClassifier())])ColumnTransformer(transformers=[('pipeline',
Pipeline(steps=[('imputer',
SimpleImputerCustom()),
('scaler', StandardScaler())]),
['age', 'fare']),
('onehotencoder',
OneHotEncoder(handle_unknown='ignore'),
['sex']),
('familyfeaturetransformer',
FamilyFeatureTransformer(),
['parch', 'sibsp'])],
verbose_feature_names_out=False)['age', 'fare']
SimpleImputerCustom()
StandardScaler()
['sex']
OneHotEncoder(handle_unknown='ignore')
['parch', 'sibsp']
FamilyFeatureTransformer()
FunctionTransformer(func=<function get_dataframe at 0x00000161953C3670>,
kw_args={'transformer': ColumnTransformer(transformers=[('pipeline',
Pipeline(steps=[('imputer',
SimpleImputerCustom()),
('scaler',
StandardScaler())]),
['age',
'fare']),
('onehotencoder',
OneHotEncoder(handle_unknown='ignore'),
['sex']),
('familyfeaturetransformer',
FamilyFeatureTransformer(),
['parch',
'sibsp'])],
verbose_feature_names_out=False)})RandomForestClassifier()
We can also get the importance of features in our dataset from RandomForrest classifier.
import matplotlib.pyplot as plt
clf = pipe[-1] # last estimator is the RF classifier
importances = clf.feature_importances_
features = clf.feature_names_in_
indices = np.argsort(importances)
plt.title('Feature Importances')
plt.barh(range(len(indices)), importances[indices], color='b', align='center')
plt.yticks(range(len(indices)), [features[i] for i in indices])
plt.xlabel('Relative Importance')
plt.show()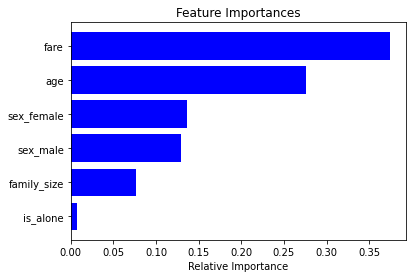
Note that all the feature names were passed to the RF Classifier and that is why we were able to get them back using its attribute feature_names_in_. This can be super useful when you have many model deployed in the environment, and you can just use the model object to get information about the features it was trained on.
For a moment let’s also remove the feature names from our pipeline and see how it will effect our feature importance plot.
pipe = Pipeline([
("preprocess", preprocessor),
# ("dataframer", dataframer),
('rf_estimator', RandomForestClassifier())
])
# fit the pipeline
pipe.fit(X_train, y_train)
# get the feature importance plot
clf = pipe[-1]
importances = clf.feature_importances_
features = clf.feature_names_in_
indices = np.argsort(importances)
plt.title('Feature Importances')
plt.barh(range(len(indices)), importances[indices], color='b', align='center')
plt.yticks(range(len(indices)), [features[i] for i in indices])
plt.xlabel('Relative Importance')
plt.show()--------------------------------------------------------------------------- AttributeError Traceback (most recent call last) Input In [38], in <module> 12 clf = pipe[-1] 13 importances = clf.feature_importances_ ---> 14 features = clf.feature_names_in_ 16 indices = np.argsort(importances) 18 plt.title('Feature Importances') AttributeError: 'RandomForestClassifier' object has no attribute 'feature_names_in_'
No feature names were passed to our classifier this time and it is missing feature_names_in_ attribute. We can circumvent this and still get feature importance plot.
pipe = Pipeline([
("preprocess", preprocessor),
# ("dataframer", dataframer),
('rf_estimator', RandomForestClassifier())
])
# fit the pipeline
pipe.fit(X_train, y_train)
# get the feature importance plot
clf = pipe[-1]
importances = clf.feature_importances_
# features = clf.feature_names_in_
indices = np.argsort(importances)
plt.title('Feature Importances')
plt.barh(range(len(indices)), importances[indices], color='b', align='center')
plt.yticks(range(len(indices)), [i for i in indices])
plt.xlabel('Relative Importance')
plt.show()
This time we get the same plot but not withOUT feature names, and it is not useful anymore. So definitely we need to keep the feature names with the final estimator. Feature names can help us a lot in interpreting the model.
For an easy reference, let’s put the whole pipeline in one place. ### Load Data
from sklearn.datasets import fetch_openml
from sklearn.model_selection import train_test_split
import pandas as pd
import numpy as np
np.random.seed(42) # for consistency
X, y = fetch_openml("titanic", version=1, as_frame=True, return_X_y=True)
X.drop(['body', 'cabin', 'boat', 'home.dest'], axis=1, inplace=True)
X_train, X_test, y_train, y_test = train_test_split(X, y, stratify=y, test_size=0.2)
X_train.head()| pclass | name | sex | age | sibsp | parch | ticket | fare | embarked | |
|---|---|---|---|---|---|---|---|---|---|
| 999 | 3.0 | McCarthy, Miss. Catherine 'Katie' | female | NaN | 0.0 | 0.0 | 383123 | 7.7500 | Q |
| 392 | 2.0 | del Carlo, Mrs. Sebastiano (Argenia Genovesi) | female | 24.0 | 1.0 | 0.0 | SC/PARIS 2167 | 27.7208 | C |
| 628 | 3.0 | Andersson, Miss. Sigrid Elisabeth | female | 11.0 | 4.0 | 2.0 | 347082 | 31.2750 | S |
| 1165 | 3.0 | Saad, Mr. Khalil | male | 25.0 | 0.0 | 0.0 | 2672 | 7.2250 | C |
| 604 | 3.0 | Abelseth, Miss. Karen Marie | female | 16.0 | 0.0 | 0.0 | 348125 | 7.6500 | S |
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import OneHotEncoder, FunctionTransformer, StandardScaler
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.compose import make_column_transformer
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score
from sklearn.impute import SimpleImputer
class FamilyFeatureTransformer(BaseEstimator, TransformerMixin):
def __init__(self):
pass
def fit(self, X, y):
return self
def transform(self, X):
X['family_size'] = X['parch'] + X['sibsp']
X.drop(['parch', 'sibsp'], axis=1, inplace=True) # we can drop this feature now
X['is_alone'] = 1
X.loc[X['family_size'] > 1, 'is_alone'] = 0
return X
def get_feature_names_out(self, input_features=None):
# this time we have created new features. Their names are different from input features.
# so we have explicitly mentioned them here.
return ['family_size', 'is_alone']
class SimpleImputerCustom(BaseEstimator, TransformerMixin):
def __init__(self, strategy='mean'):
self.strategy = strategy
self.imputer = SimpleImputer(strategy=self.strategy)
def fit(self, X, y):
self.imputer.fit(X, y)
return self
def transform(self, X):
return self.imputer.transform(X)
def get_feature_names_out(self, input_features=None):
# we have returned the input features name as out features will have the same name
return input_features
def get_dataframe(X, transformer):
"""
x: an nd-array
transformer: fitted transformer
"""
col_names = transformer.get_feature_names_out()
output = pd.DataFrame(X.T, col_names).T
return output
numeric_features = ["age", "fare"]
numeric_transformer = Pipeline(
steps=[
("imputer", SimpleImputerCustom(strategy='mean')),
("scaler", StandardScaler())
]
)
categorical_features = ["sex"]
categorical_transformer = OneHotEncoder(handle_unknown="ignore")
family_features = ["parch", "sibsp"]
family_transformer = FamilyFeatureTransformer()
preprocessor = make_column_transformer(
(numeric_transformer, numeric_features),
(categorical_transformer, categorical_features),
(family_transformer, family_features),
remainder='drop', # let's drop extra features this time
verbose_feature_names_out=False
)
dataframer = FunctionTransformer(func=get_dataframe, kw_args={"transformer": preprocessor})
pipe = Pipeline([
("preprocess", preprocessor),
("dataframer", dataframer),
('rf_estimator', RandomForestClassifier())
])
pipe.fit(X_train, y_train)
y_pred = pipe.predict(X_test)
accuracy_score(y_test, y_pred)0.8015267175572519import matplotlib.pyplot as plt
clf = pipe[-1] # last estimator is the RF classifier
importances = clf.feature_importances_
features = clf.feature_names_in_
indices = np.argsort(importances)
plt.title('Feature Importances')
plt.barh(range(len(indices)), importances[indices], color='b', align='center')
plt.yticks(range(len(indices)), [features[i] for i in indices])
plt.xlabel('Relative Importance')
plt.show()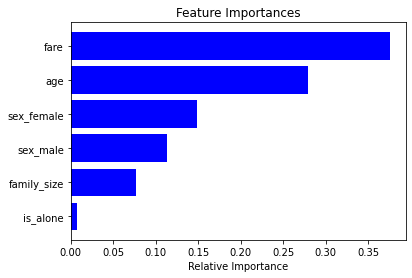
from sklearn import set_config
set_config(display="diagram")
import pickle
# save our pipeline
s1 = pickle.dumps(pipe)
# reload it
s2 = pickle.loads(s1)
s2Pipeline(steps=[('preprocess',
ColumnTransformer(transformers=[('pipeline',
Pipeline(steps=[('imputer',
SimpleImputerCustom()),
('scaler',
StandardScaler())]),
['age', 'fare']),
('onehotencoder',
OneHotEncoder(handle_unknown='ignore'),
['sex']),
('familyfeaturetransformer',
FamilyFeatureTransformer(),
['parch', 'sibsp'])],
verbose_feature_names_out=False)),
('data...
kw_args={'transformer': ColumnTransformer(transformers=[('pipeline',
Pipeline(steps=[('imputer',
SimpleImputerCustom()),
('scaler',
StandardScaler())]),
['age',
'fare']),
('onehotencoder',
OneHotEncoder(handle_unknown='ignore'),
['sex']),
('familyfeaturetransformer',
FamilyFeatureTransformer(),
['parch',
'sibsp'])],
verbose_feature_names_out=False)})),
('rf_estimator', RandomForestClassifier())])Please rerun this cell to show the HTML repr or trust the notebook.Pipeline(steps=[('preprocess',
ColumnTransformer(transformers=[('pipeline',
Pipeline(steps=[('imputer',
SimpleImputerCustom()),
('scaler',
StandardScaler())]),
['age', 'fare']),
('onehotencoder',
OneHotEncoder(handle_unknown='ignore'),
['sex']),
('familyfeaturetransformer',
FamilyFeatureTransformer(),
['parch', 'sibsp'])],
verbose_feature_names_out=False)),
('data...
kw_args={'transformer': ColumnTransformer(transformers=[('pipeline',
Pipeline(steps=[('imputer',
SimpleImputerCustom()),
('scaler',
StandardScaler())]),
['age',
'fare']),
('onehotencoder',
OneHotEncoder(handle_unknown='ignore'),
['sex']),
('familyfeaturetransformer',
FamilyFeatureTransformer(),
['parch',
'sibsp'])],
verbose_feature_names_out=False)})),
('rf_estimator', RandomForestClassifier())])ColumnTransformer(transformers=[('pipeline',
Pipeline(steps=[('imputer',
SimpleImputerCustom()),
('scaler', StandardScaler())]),
['age', 'fare']),
('onehotencoder',
OneHotEncoder(handle_unknown='ignore'),
['sex']),
('familyfeaturetransformer',
FamilyFeatureTransformer(),
['parch', 'sibsp'])],
verbose_feature_names_out=False)['age', 'fare']
SimpleImputerCustom()
StandardScaler()
['sex']
OneHotEncoder(handle_unknown='ignore')
['parch', 'sibsp']
FamilyFeatureTransformer()
FunctionTransformer(func=<function get_dataframe at 0x00000161997DF3A0>,
kw_args={'transformer': ColumnTransformer(transformers=[('pipeline',
Pipeline(steps=[('imputer',
SimpleImputerCustom()),
('scaler',
StandardScaler())]),
['age',
'fare']),
('onehotencoder',
OneHotEncoder(handle_unknown='ignore'),
['sex']),
('familyfeaturetransformer',
FamilyFeatureTransformer(),
['parch',
'sibsp'])],
verbose_feature_names_out=False)})RandomForestClassifier()